The best place i found for learning smiles format specs is
http://www.daylight.com/dayhtml/doc/theory/theory.smiles.htmlA smiles file consist just in a text file with a single line which specifies chemical composition and structure. In the table below some examples are shown.
| molecule name | smiles specification |
|---|---|
| methanol | CO |
| ammonium | [NH4+] |
| D-glucose | O=C[C@H](O)[C@@H](O)[C@H](O)[C@H](O)C(O) |
| L-proline | [O-]C(=O)C1CCC[NH2+]1 |
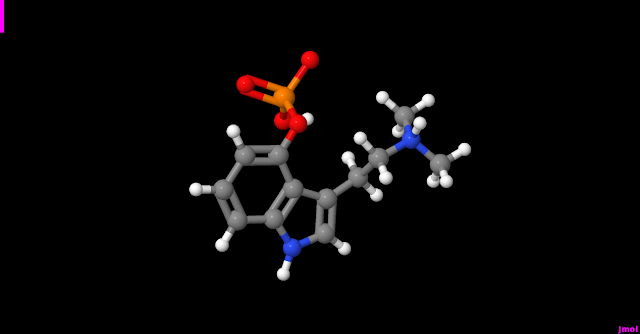
| psilocybin | O(P(O)(=O)[O-])c1cccc2c1c(CC[NH+](C)C)cn2 |
| iron(III) oxide | O=[Fe]O[Fe]=O |
| Sodium chloride | [Na+].[Cl-] |
Another great advantage of using this approach is that most chemical compounds found in wikipedia comes with the smiles specification string.
Once you have the smiles file with the molecule spec, you can easily convert to jmol format file (.mol extension), using babel:
The --gen3d option is needed in order to generate 3D coordinates in the destination file. babel will also perform energy minimization calculations in order to create a valid molecule configuration.linux$ babel myfile.smi myfile.mol --gen3d
Finally, you open it with jmol:
The image is an example on how a molecule looks in the jmol visualizer area (jmol can export a view as an image in different formats), according to ball-and-stick model:linux$ jmol myfile.mol
Once in the jmol visualizer, you can move and rotate the molecule easily with the mouse:
- rotate on X axis: press left button and move the mouse pointer up/down without releasing the button.
- rotate on Y axis: press left button and move the mouse pointer left/right without releasing the button.
- rotate on Z axis: press SHIFT key, then right button, and move the mouse pointer left/right without releasing the key nor the button.
- move away/closer: use the mouse wheel.
- translate: double click left button and move mouse pointer without releasing button.
Other interesting functions are accesible by clicking right mouse on the visualization area. For example, with menu View you can change the perspective from the six sides of the molecule bounding box. Also you can active spin on and off in order to view the molecule from a rotating camera around the object.
Jmol commands are also accesible from a console (to open: Menu File -> Console) and in fact advanced control and computations can only be done from the console.
For instance, spin can be set on/off from the console using the jmol command spin. Examples:
jmol$ spin on
In order to reset the molecule to its initial position:jmol$ spin off
When you place the mouse over an atom, a label will be displayed (by default, the element symbol plus the number id of the atom, but can be controlled with the command hover). You can use this label with the command center in order to center the molecule at the given atom. Rotation operations then will be centered on the selected atom. Examples:jmol$ reset
jmol$ center C3
jmol$ center N11
A complete reference of jmol commands and functions can be found here:
http://chemapps.stolaf.edu/jmol/docs/
No comments:
Post a Comment